# init repo notebook
!git clone https://github.com/rramosp/ppdl.git > /dev/null 2> /dev/null
!mv -n ppdl/content/init.py ppdl/content/local . 2> /dev/null
!pip install -r ppdl/content/requirements.txt > /dev/null
Bayes theorem continuous#
import numpy as np
import matplotlib.pyplot as plt
from scipy import stats
from scipy.integrate import quad
from rlxutils import subplots, copy_func
from progressbar import progressbar as pbar
%matplotlib inline
def get_posterior(likelihood_fn, prior_fn, support, atol=1e-5, integration_intervals=1000):
assert len(support)==2
xr = np.linspace(*support, integration_intervals)
Z = np.sum([likelihood_fn(x)*prior_fn(x) for x in xr]) * (xr[1]-xr[0])
posterior_fn = lambda x: copy_func(likelihood_fn)(x) * copy_func(prior_fn)(x) / Z
return posterior_fn
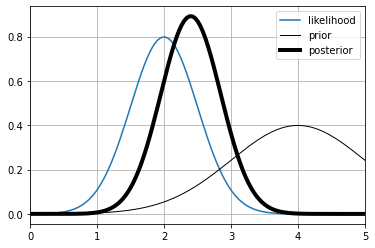
the posterior of one measurement#
support = [-20,20]
likelihood = lambda x: stats.norm(loc=2,scale=.5).pdf(x)
prior = lambda x: stats.norm(loc=4,scale=1).pdf(x)
posterior = get_posterior(likelihood, prior, support)
xr = np.linspace(*support,1000)
plt.plot(xr, likelihood(xr), label="likelihood")
plt.plot(xr, prior(xr), label="prior", color="black", lw=1)
plt.plot(xr, posterior(xr), label="posterior", color="black", lw=4)
plt.grid();
plt.legend();
plt.xlim(0,5);
several measurements#
we repeat the steps above, using the posterior obtained at each step as the prior for next measurement
def estimate_position(
z_real = 23,
prior_mean = 20,
prior_std = 10,
sensor_sigma = .7,
support = [-10, 50]
):
initial_prior = eval(f"lambda x: stats.norm(loc={prior_mean},scale={prior_std}).pdf(x)")
measurements = np.random.normal(loc=z_real, scale=sensor_sigma, size=10)
posteriors = []
prior = initial_prior
for i in pbar(range(len(measurements))):
# compute the likelihood
likelihood = eval(f"lambda x: stats.norm(loc={measurements[i]},scale={sensor_sigma}).pdf(x)")
# compute the posterior, stop if integration errors
posterior = get_posterior(likelihood, prior, support)
posteriors.append(posterior)
# the posterior becomes the next prior
prior = posteriors[-1]
# plot stuff
xr = np.linspace(*support, 10000)
for ax,i in subplots(len(posteriors)+1, n_cols=6):
if i==0:
plt.plot(xr, initial_prior(xr), color="steelblue")
plt.title("prior")
plt.ylim(0,0.5)
plt.xlim(z_real-5, z_real+5)
else:
plt.plot(xr, posteriors[i-1](xr), color="steelblue")
plt.title(f"measurement {i}")
plt.axvline(measurements[i-1], color="black", ls="--")
plt.xlim(z_real-2, z_real+2)
plt.grid();
plt.axvline(z_real, color="black")
plt.tight_layout()
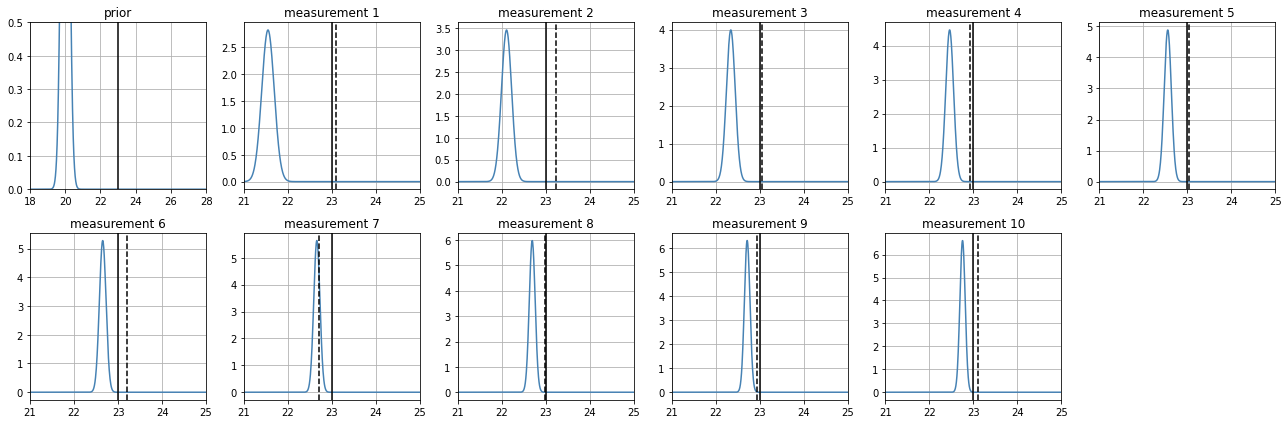
a very uninformative prior#
observe how, as measurements are processed, the posterior becomes narrower (less uncertainty) and approaches the real value
estimate_position(
z_real = 23,
prior_mean = 5,
prior_std = 20,
sensor_sigma = .7
)
100% (10 of 10) |########################| Elapsed Time: 0:00:27 Time: 0:00:27
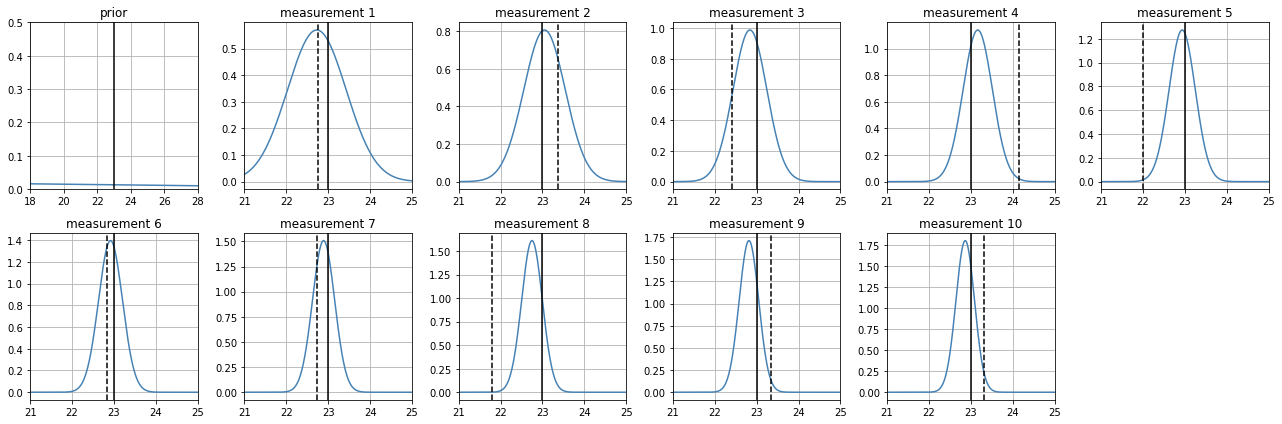
a more accurate sensor converges faster and with less ucertainty#
however observe that numerical integration might stop us from processing further measurements
estimate_position(
z_real = 23,
prior_mean = 20,
prior_std = 10,
sensor_sigma = .2
)
100% (10 of 10) |########################| Elapsed Time: 0:00:28 Time: 0:00:28
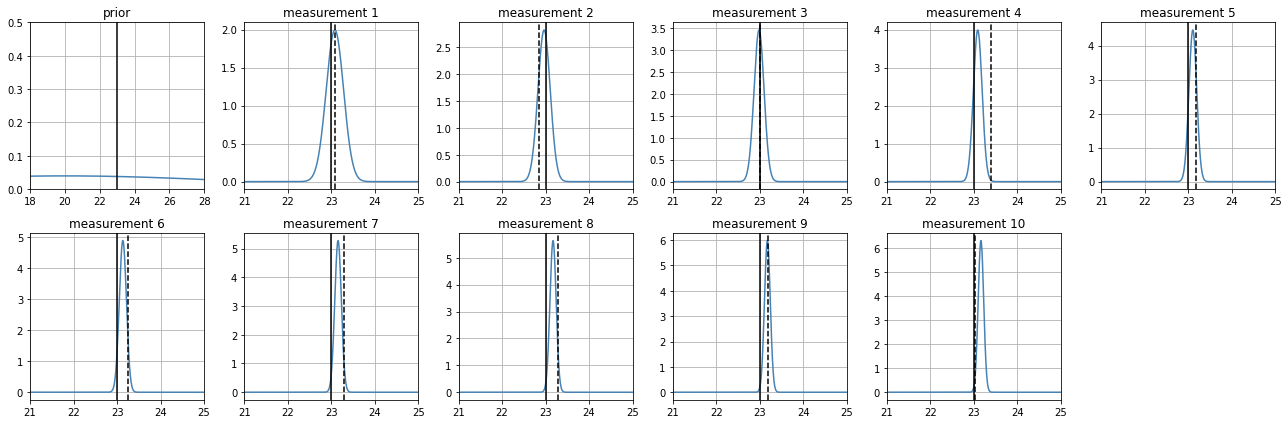
but if we are very wrong and very confident on our prior, we pay for it#
Wrong mean with a very narrow stdev. It takes many measurements to converge to the real value.
Recall, narrow stdevs anywhere always represent small uncertainty
estimate_position(
z_real = 23,
prior_mean = 20,
prior_std = .2,
sensor_sigma = .2
)
100% (10 of 10) |########################| Elapsed Time: 0:00:28 Time: 0:00:28